Marat Khamadeev
Marat Khamadeev

Chemical language models demonstrated a poor understanding of chemistry
The application of language models in chemistry (ChemLMs) has opened up new opportunities for the automatic analysis of substance properties, the generation of new compounds, and the prediction of reactions. For this purpose, the molecules, reaction equations, and other chemical information must be transformed into a textual format. Molecules can be represented in formats like SMILES strings, IUPAC names, and others. Cross-domain models are also trained on the task of molecule captioning to describe molecules in a natural language.
Several modern models, such as ChemBERTa or MolT5, are actively being developed based on this principle. These models demonstrate decent performance on the downstream chemical tasks. However, the question remains open whether modern language models have truly learned to understand the basic laws of chemistry or if they are merely memorizing molecule representations.
A team of NLP researchers from AIRI and Sber decided to address this question by creating AMORE — a new and flexible zero-shot framework that assesses the trustworthiness of ChemLMs. It is suitable for models trained solely on molecules for chemical tasks and on a combined corpus of natural language texts and string-based structures.
At the core of AMORE framework lies the similarity between the embedding representations of the molecule, its SMILES variation, and that of another molecule. The authors selected four augmentations: canonicalization, explicit addition of hydrogen atoms in the notation, kekulization, and cycle renumbering. All these modifications change the string representation but denote the same chemical substance. The researchers conducted experiments on two popular datasets: the ChEBI-20 dataset, which contains about 3,000 molecule-description pairs, and an isomeric molecules subset from the QM9 dataset. Descriptions of isomeric molecules were found by authors in the PubChem database.
The evaluation showed that existing ChemLMs are not robust even to simple augmentations of SMILES strings, and the quality of generated molecule descriptions significantly decreased on the augmented data. Furthermore it was found during the experiments that the AMORE score changes similarly to classical text generation metrics such as ROUGE and METEOR.
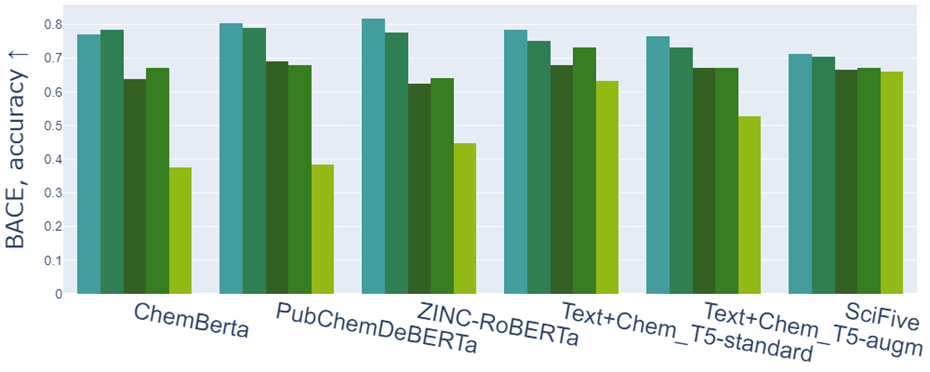
The drop in accuracy of some models on the task of classifying molecules based on their ability to inhibit human β-secretase (BACE 1 task). The colors from left to right encode the original data and their augmentations: cycle renumbering, canonicalization, kekulization, and hydrogen addition.
The results of the researchers' work demonstrate that the existing ChemLMs have problems with distinguishing the same molecules in different representations. Thus, there is currently no satisfactory understanding of the laws of chemistry in modern chemical language models.
The article describing the method was published in the proceedings of the EMNLP 2024 conference, and the code is open and available on GitHub.