Marat Khamadeev
Marat Khamadeev

Language models for chemical tasks turned out to be vulnerable to changes in molecule representations
To predict the outcome of a chemical reaction, one can describe it as the interaction of a multitude of neutral atoms, ions, and electrons, having a quantum nature. However, in quantum mechanics, only the two-body problem (hydrogen-like atoms or particle scattering) is precisely solved. To predict, for example, the properties of a helium atom, approximate or numerical methods are required. As the number of particles increases, the complexity of calculations significantly rises, making traditional quantum chemistry methods very demanding on computational resources.
Addressing these challenges may involve the application of various machine-learning approaches that attempt to provide answers by generalizing chemical laws based on extensive datasets. For instance, we have already discussed how graph neural networks are used for this purpose. Atoms are located at their nodes, and edges correspond to chemical bonds.
Another approach to this problem is based on the use of transformer architecture, which is specifically designed for processing long textual sequences. In this case, the molecule is represented as a sequence of symbols using the SMILES language. This allows feeding language models huge amounts of data on molecules and their properties and then attempting to solve various tasks: generating new compounds, predicting physicochemical properties, describing reactions, and so on.
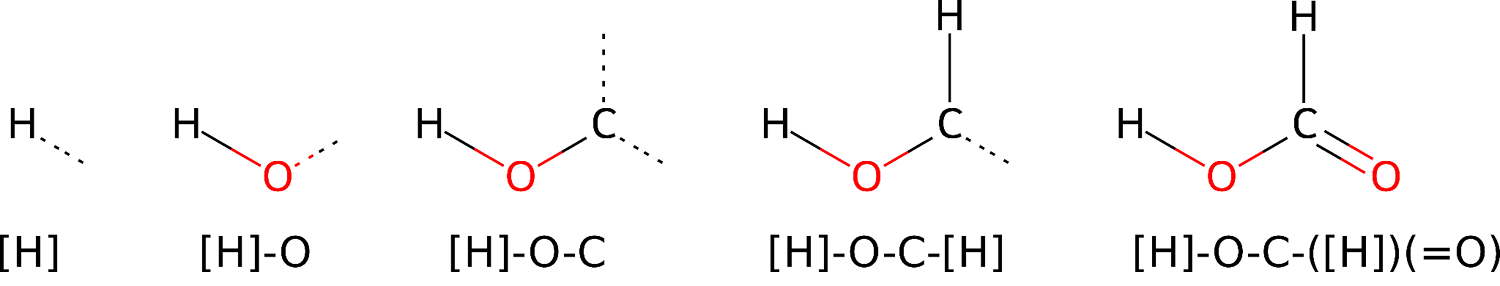
Building the SMILES representation of formic acid. Source: Maria Kadukova / biomolecula.ru
The next step in developing this paradigm is the creation of cross-domain language models that learn to link chemical data with words that describe them. To test them, researchers are developing various benchmarks and tests that allow evaluating the models' ability to retain correct chemical knowledge. One such test was developed by a team of researchers led by Elena Tutubalina, who heads the "Domain-specific NLP" group at AIRI.
The authors set for the models a series of tasks involving the text description of molecules and their properties, which are similar to translating SMILES text into human language. They evaluated the models' understanding of chemistry using two popular examples in this field: MolT5 and Text+Chem T5, each with two versions.
During the experiments, researchers found that these models remain vulnerable to even slight changes in the symbolic molecule representations, even though the notation remains a correct representation of the same molecule from a chemistry perspective. They were able to demonstrate that such changes led to a decrease in output quality and accuracy. Still, the extent of this reduction seems to be mostly dictated by language processing rather than an underlying understanding of chemistry. This research will help better understand the weaknesses not only in cross-modal models in the field of chemistry but also in language cross-domain models in general.
The work was presented at the ICLR 2024 conference, and the article was published in its proceedings. The source code is available on GitHub.